A Comparison of Common Regression Ensembles¶
On the following page I train and compare common types of neural network regression ensembles: Mixtures of Experts, Sparsely Gated Mixtures of Experts, and Negative Correlation Learning. Additionally, I test altering the output activation on Mixtures of Experts models and test the effects of pretraining expert models.
| Model | # Base Models | Sparse Gates (if applicable) | MSE |
|---|---|---|---|
| Single Neural Network | 1 | n/a | 0.03624 |
| Mixtures of Experts | 4 | n/a | 0.017827 |
| Negative Correlation Learning, lambda=0.5 | 4 | n/a | 0.0354 |
| Negative Correlation Learning, lambda=0.1 | 4 | n/a | 0.0355601 |
| Sparse MoE | 4 | 1 | 0.02036 |
| Sparse MoE | 4 | 2 | 0.023 |
| Sparse MoE | 8 | 2 | 0.02219 |
| Sparse MoE | 8 | 4 | 0.017976 |
| Mixture of Experts, pretrained | 4 | n/a | 0.01444 |
| Mixture of Experts, pretrained with elu gating activation | 4 | n/a | 0.0010685 |
| Sparse MoE , pretrained | 8 | 2 | 0.00907 |
| Sparse MoE , pretrained | 8 | 4 | 0.00049734 |
| Sparse MoE , pretrained with elu gating activation | 8 | 2 | 0.004674 |
| Sparse MoE , pretrained with elu gating activation | 8 | 4 | 0.001323 |
**All neural networks had one hidden node. Mixture of Experts gating networks had two hidden nodes. Training was for 100 epochs with SGD optimization with a learning rate of 1.
Conclusions¶
- Pretraining expert networks with distinct subspaces performs better in every case.
- Using alternative gating activation with pretrained experts networks shows promising results.
- Sparse gating shows superior results when expert networks are pretrained.
Data¶
Data is based off the following equation:
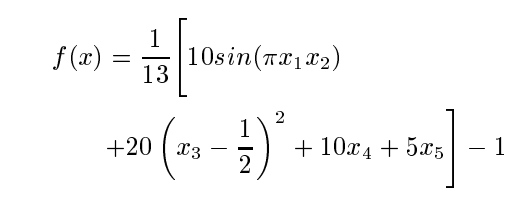
where x = [x1,... x5] is an input vetor whose components lie between zero and one. The value of f (x) lies between 1 and +1. The data onsisted of input/output patterns with the input vetors sampled uniformly at random from the interval (0,1). Training pattern set size was 500, testing set size was 10,000.
Negative Correlation Learning¶
Equations attained from http://citeseerx.ist.psu.edu/viewdoc/download;jsessionid=7E1378BAA5112F9D4775D09B977E80C0?doi=10.1.1.11.1126&rep=rep1&type=pdf
To diversify the ensemble and encourage negatively correlated submodels in an ensemble, the following loss function was implemented:
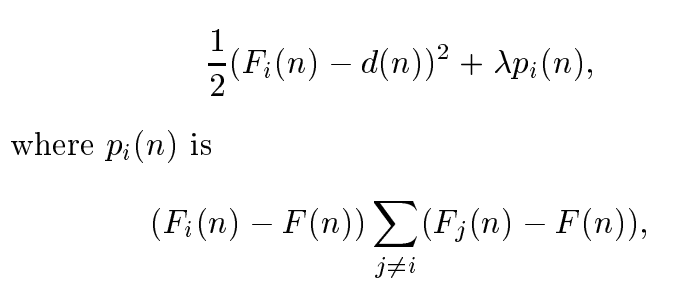
Mixtures of Experts¶
Equations attained from https://www.cs.toronto.edu/~hinton/absps/Outrageously.pdf
Loss¶
When end-to-end training mixtures of experts, diversity is encouraged using an additional term in the loss function, the coefficient of variation of the "importance". Importance is equal to the "the batchwise sum of the gate values for that expert." The loss added is equal to the square of the coefficient of variation of the set of importance values, multiplied by a hand-tuned scaling factor importance."
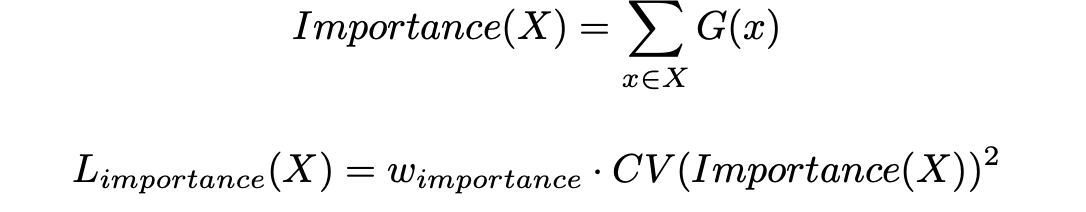
Sparse Gating¶
Before taking the softmax of the gating network, we keep only the top-k values. We set the rest of the values to -infinity because the softmax of -infinity is 0.
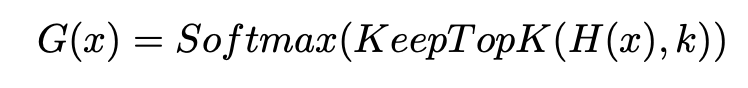 Keras/TensorFlow code for this is below.
Keras/TensorFlow code for this is below.
Main methods and parameter definitions¶
import random
import math
import numpy as np
from keras.layers import Dense,Input,Lambda,Activation
import keras
from keras.models import Model
from keras import backend as K
import sys
import os
from keras.optimizers import SGD, Adam
from keras.callbacks import ModelCheckpoint, History
from matplotlib import pyplot as plt
import matplotlib.pyplot as plt
import matplotlib
import tensorflow as tf
from sklearn.preprocessing import MinMaxScaler
#!{sys.executable} -m pip install sklearn
#!{sys.executable} -m pip install keras
def generate_input():
return [random.uniform(0, 1),random.uniform(0, 1),random.uniform(0, 1),random.uniform(0, 1),random.uniform(0, 1)]
def generate_output(inputs):
x= (1/13)*(10*math.sin(math.pi*inputs[0]*inputs[1])+20*(inputs[2]-.5)**2+10*inputs[3]+5*inputs[4])-1
return x
def generate_data(num_to_generate):
inputs=[]
outputs=[]
for a in range(num_to_generate):
x=generate_input()
y=generate_output(x)
inputs.append(x)
outputs.append(y)
return np.array(inputs), np.array(outputs)
def getData(test=False):
if(test):
#test data
if(os.path.isfile('test_in.npy') and os.path.isfile('test_out.npy')):
inputs=np.load('test_in.npy')
outputs=np.load('test_out.npy')
else:
inputs,outputs=generate_data(10000)
np.save('test_in.npy', inputs)
np.save('test_out.npy',outputs)
else:
#train data
if(os.path.isfile('train_in.npy') and os.path.isfile('train_out.npy')):
inputs=np.load('train_in.npy')
outputs=np.load('train_out.npy')
else:
inputs,outputs=generate_data(500)
np.save('train_in.npy', inputs)
np.save('train_out.npy',outputs)
return inputs, outputs
def nn(num,H):
nn=Dense(H,kernel_initializer='random_uniform', name='base1_'+str(num))(nn_inputs)
nn=Dense(1,name='base2_'+str(num),kernel_initializer='random_uniform')(nn)
return Model(inputs=nn_inputs, outputs=nn)
#Tried different weight initializations
#weight_init=keras.initializers.RandomUniform(minval = 0, maxval = 0.05)
#weight_init=keras.initializers.Zeros()
#weight_init=keras.initializers.RandomNormal(mean=0.1, stddev=0.05, seed=None)
#,kernel_initializer=weight_init
def moe_(H, num_models):
nn=Dense(H, name='hiddenlayer1')(nn_inputs)
#nn=Activation('sigmoid', name='hiddenactivation')(nn)
nn=Dense(num_models,name='gate')(nn)
return Model(inputs=nn_inputs, outputs=nn)
def ensemble_average(branches):
forLambda=[]
forLambda.extend(branches)
add= Lambda(lambda x:K.tf.transpose(sum(K.tf.transpose(forLambda[i]) for i in range(0,len(forLambda)))/len(forLambda)), name='final')(forLambda)
return add
#MOE
def gating_multiplier(gate,branches):
forLambda=[gate]
forLambda.extend(branches)
add= Lambda(lambda x:K.tf.transpose(
sum(K.tf.transpose(forLambda[i]) *
forLambda[0][:, i-1] for i in range(1,len(forLambda))
)
))(forLambda)
return add
def slices_to_dims(slice_indices):
"""
Args:
slice_indices: An [N, k] Tensor mapping to column indices.
Returns:
An index Tensor with shape [N * k, 2], corresponding to indices suitable for
passing to SparseTensor.
"""
slice_indices = tf.cast(slice_indices, tf.int64)
num_rows = tf.shape(slice_indices, out_type=tf.int64)[0]
row_range = tf.range(num_rows)
item_numbers = slice_indices * num_rows + tf.expand_dims(row_range, axis=1)
item_numbers_flat = tf.reshape(item_numbers, [-1])
return tf.stack([item_numbers_flat % num_rows,
item_numbers_flat // num_rows], axis=1)
def plot(model):
figure = plt.figure(figsize=(18, 16))
X = np.arange(0,len(outputs))
tick_plot = figure.add_subplot(1, 1, 1)
tick_plot.plot(X,outputs, color='green', linestyle='-', marker='*', label='Actual')
tick_plot.plot(X, model.predict(inputs), color='orange',linestyle='-', marker='*', label='Predictions')
plt.xlabel('')
plt.ylabel('')
plt.legend(loc='upper left')
error=model.evaluate(inputs,outputs)
plt.title('Total MSE: '+str(error))
print("Ensemble MSE's:")
print(errors)
plt.show()
def sub_model_errors(model):
errors=[]
test_x, test_y=getData(test=True)
#print(outputs[:1])
for i in range(2,num_models+2):
model1=Model(inputs=nn_inputs, outputs=model.layers[-i].output)
model1.compile(loss='mse', optimizer=SGD(lr=0.1))
errors.append(model1.evaluate(test_x, test_y))
return errors
def cv_squared(x):
"""The squared coefficient of variation of a sample.
Useful as a loss to encourage a positive distribution to be more uniform.
Epsilons added for numerical stability.
Returns 0 for an empty Tensor.
Args:
x: a `Tensor`.
Returns:
a `Scalar`.
"""
epsilon = 1e-10
float_size = tf.to_float(tf.size(x)) + epsilon
mean = tf.reduce_sum(x) / float_size
variance = tf.reduce_sum(tf.squared_difference(x, mean)) / float_size
return variance / (tf.square(mean) + epsilon)
def sparseGating(inputs_,gates=2):
indi=tf.cast(tf.math.top_k(inputs_,gates, sorted=False).indices,dtype=tf.int64)
v=tf.math.top_k(inputs_,gates, sorted=False).values
sparse_indices = slices_to_dims(indi)
sparse = tf.SparseTensor( indices=sparse_indices, values=tf.reshape(v, [-1]),
dense_shape=tf.cast(tf.shape(inputs_),dtype=tf.int64))
c=tf.zeros_like(inputs_)
d=tf.sparse_add(c, sparse)
z =tf.ones_like(inputs_)*-np.inf
mask = tf.less_equal(d, tf.zeros_like(d))
new_tensor = tf.multiply(z, tf.cast(mask, dtype=tf.float32))
g=tf.where(tf.is_nan(new_tensor), tf.zeros_like(new_tensor), new_tensor)
g=tf.sparse_add(g,sparse)
b=Lambda(lambda a:g)(inputs_)
return b
def preTrain(weight_file):
maxVal=np.amax(train_y)
minVal=np.amin(train_y)
diff=maxVal-minVal
for i in range(num_models):
test_val_min=diff/num_models*i+minVal
test_val_max=diff/num_models*(i+1)+minVal
y=[j for j in range(len(train_y)) if train_y[j]>test_val_min and train_y[j]<test_val_max]
x=train_x[y]
y=train_y[y]
models[i].compile(loss='mse', optimizer=SGD(lr=0.1))
file=str(weight_file)+'_'+str(i)+'.h5'
checkpointer=ModelCheckpoint(file, monitor='loss', verbose=1, save_best_only=True, save_weights_only=True, mode='auto')
models[i].fit(x,y,epochs=100, verbose=1,batch_size=1,callbacks=[checkpointer])
def load_weights(model,weight_file):
for a in range(num_models):
m=models[a]
file=str(weight_file)+'_'+str(a)+'.h5'
m.load_weights(file,by_name=True)
for b in m.layers:
for l in model.layers:
if(l.name==b.name):
l.set_weights(b.get_weights())
print("Loaded: "+l.name)
for l in model.layers:
if('base' in l.name):
l.trainable=True
else:
print(l.name)
def moeLoss(yTrue,yPred):
loss_calc=0
j=0
importance=[]
for i in range(4):
importance.append(tf.reduce_sum(model.get_layer('gate').output[:,i]))
for i in reversed(range(2,num_models+2)):
loss_calc+=(model.get_layer('act').output[:,j]*tf.math.exp(-1/2*(yTrue-model.layers[-i].output)**2))
j+=1
loss_calc=-tf.math.log(loss_calc)
return (loss_calc+.1*cv_squared(importance))
def negativeCorrelation(yTrue,yPred):
#return K.mean(K.square(yPred - yTrue), axis=-1)
loss_calc=0
for i in range(2,num_models+2):
others=0
for j in range (2,num_models+2):
if(j==i):
continue
else:
others+=(model.layers[-j].output-model.layers[-1].output)
loss_calc+=1/2*(model.layers[-i].output-yTrue)**2+(lambda_*(model.layers[-i].output-model.layers[-1].output)*(others))
return loss_calc
Model definitions¶
model_to_run='neg_cor'
if(model_to_run=='single_nn'):
#Best run on test set MSE: 0.03624
model=nn("x", H)
model.compile(loss='mse', optimizer=SGD(lr=0.1))
model_single_nn=model
checkpointer=ModelCheckpoint('single.h5', monitor='loss', verbose=1, save_best_only=True, save_weights_only=True, mode='auto')
elif(model_to_run=='moe'):
#Best run on test set MSE: 0.017827
num_models=4
models=[nn(i, H) for i in range(num_models)]
moe=moe_(expert_nodes, num_models)
layer=Activation("softmax",name='act')(moe.output)
gate=gating_multiplier(layer,[m.layers[-1].output for m in models])
model=Model(inputs=nn_inputs, outputs=gate)
model.compile(loss=moeLoss, optimizer=SGD(lr=0.1))
checkpointer=ModelCheckpoint('moe.h5', monitor='loss', verbose=1, save_best_only=True, save_weights_only=True, mode='auto')
elif(model_to_run=='neg_cor'):
num_models=4
lambda_=0.1
models=[nn(i, H) for i in range(num_models)]
model_out=ensemble_average([models[0].output,models[1].output,models[2].output,models[3].output])
model=Model(inputs=nn_inputs, outputs=model_out)
model.compile(loss=negativeCorrelation, optimizer=SGD(lr=0.1))
checkpointer=ModelCheckpoint('averages.h5', monitor='loss', verbose=1, save_best_only=True, save_weights_only=True, mode='auto')
elif(model_to_run=='sparse'):
#Best run on test set MSE models=4, gates=1: 0.02036
#Best run on test set MSE models=4, gates=2: 0.023095
#Best run on test set MSE models=8, gates=2: 0.02219
#Best run on test set MSE models=8, gates=4: 0.017976
num_models=8
models=[nn(i, H) for i in range(num_models)]
num_gates=4
moe=moe_(expert_nodes, num_models)
sparse_layer=sparseGating(moe.output, gates=num_gates)
layer=Activation("softmax",name='act')(sparse_layer)
layer=gating_multiplier(layer,[m.layers[-1].output for m in models])
model=Model(inputs=nn_inputs, outputs=layer)
checkpointer=ModelCheckpoint('sparse.h5', monitor='loss', verbose=1, save_best_only=True, save_weights_only=True, mode='auto')
model.compile(loss=moeLoss, optimizer=SGD(lr=0.1))
elif(model_to_run=='moe_pretrained_elu'):
#Best run on test set MSE: 0.0010685
num_models=4
models=[nn(i, H) for i in range(num_models)]
#preTrain()
moe=moe_(expert_nodes, num_models)
layer=Activation("elu",name='act')(moe.output)
layer=gating_multiplier(layer,[m.layers[-1].output for m in models])
model=Model(inputs=nn_inputs, outputs=layer)
load_weights(model)
checkpointer=ModelCheckpoint('moe_pretrained_elu.h5', monitor='loss', verbose=1, save_best_only=True, save_weights_only=True, mode='auto')
model.compile(loss='mse', optimizer=SGD(lr=0.1))
elif(model_to_run=='moe_pretrained'):
#Best run on test set MSE: 0.01444
num_models=4
models=[nn(i, H) for i in range(num_models)]
preTrain('base1')
moe=moe_(expert_nodes, num_models)
layer=gating_multiplier(moe.output,[m.layers[-1].output for m in models])
model=Model(inputs=nn_inputs, outputs=layer)
load_weights(model,'base1')
checkpointer=ModelCheckpoint('moe_pretrained.h5', monitor='loss', verbose=1, save_best_only=True, save_weights_only=True, mode='auto')
model.compile(loss='mse', optimizer=SGD(lr=0.1))
elif(model_to_run=='sparse_pretrained'):
#Best run on test set MSE models=8, gates=2: 0.00907
#Best run on test set MSE models=8, gates=4: 0.00049734
num_models=8
models=[nn(i, H) for i in range(num_models)]
preTrain('base1')
moe=moe_(expert_nodes, num_models)
num_gates=4
layer=sparseGating(moe.output, gates=num_gates)
layer=Activation("softmax",name='act')(layer)
layer=gating_multiplier(layer,[m.layers[-1].output for m in models])
model=Model(inputs=nn_inputs, outputs=layer)
load_weights(model,'base1')
checkpointer=ModelCheckpoint('sparse_pretrained.h5', monitor='loss', verbose=1, save_best_only=True, save_weights_only=True, mode='auto')
model.load_weights('sparse_pretrained.h5')
model.compile(loss='mse', optimizer=SGD(lr=0.1))
elif(model_to_run=='sparse_pretrained_elu'):
#Best run on test set MSE models=8, gates=2: 0.004674
#Best run on test set MSE models=8, gates=4: 0.001323
num_models=8
models=[nn(i, H) for i in range(num_models)]
preTrain('base1')
moe=moe_(expert_nodes, num_models)
num_gates=4
layer=Activation("elu",name='act2')(moe.output)
layer=sparseGating(layer, gates=num_gates)
layer=Activation("softmax",name='act')(layer)
layer=gating_multiplier(layer,[m.layers[-1].output for m in models])
model=Model(inputs=nn_inputs, outputs=layer)
load_weights(model,'base1')
checkpointer=ModelCheckpoint('sparse_pretrained_elu.h5', monitor='loss', verbose=1, save_best_only=True, save_weights_only=True, mode='auto')
model.compile(loss='mse', optimizer=SGD(lr=0.1))
model.fit(train_x,train_y,epochs=100, verbose=1,batch_size=1,callbacks=[checkpointer])